Thomas G. Brooks
- Research Associate
- ITMAT Bioinformatics Lab
- Institute for Translational Medicine and Therapeutics
- University of Pennsylvania
I'm a research associate at the ITMAT Bioinformatics lab. I study transcriptomics and RNA-seq methodology, circadian biology and diurnal rhythms. Previously, I studied Riemannian geometry.
Research Interests
- Transcriptomics, RNA-seq
- Diurnal rhythms
- Circadian biology
- Methods development and benchmarking
Education
- PostDoc in Bioinformatics, 2018-2023. University of Pennsylvania
- PhD in mathematics, 2013-2018. University of Pennsylvania
- BA in mathematics, 2009-2013. Cornell University
Projects
dependentsimr
R package to simulate omics data with realistic correlation.
CorrReg
Python package to regress correlation of two variables as a function of independent variables.
Ribo Remover
Utility to remove mammalian ribosomal RNA content from FASTQ files.
See more projects on my github page.
Publications & Preprints
Filter by:
Reset
- Role of IL-10 signaling in the circadian control of host response to influenza infection Forrest KM, Towers ME, Paul O, Cho H, Padmini MV, Naik A, Lim HEK, Issah Y, Tang SY, Deshmukh HS, Abt MC, Hudock KM, Eisenlohr LC, Grant GR, Brooks TG, Sengupta S. Mucosal Immunology. 2025 November.
- Critically ill patients with a reverse blood pressure dipping phenotype at increased risk for delirium and death El Jamal N, Brooks TG, Mrcela A, Genuardi MV, FitzGerald GA, Skarke C. Scientific Reports. 2025 November.
- Effect of external cues on clock-driven protection from influenza A infection Paul O, Brooks TG, Shetty A, Choi YJ, Towers M, Assi LJ, Garifallou JP, Forrest K, Cameron A, Sehgal A, Grant G, Sengupta S. JCI. 2025 November.
- Circadian control of pulmonary endothelial signaling occurs via the NADPH oxidase 2-NLRP3 pathway Sengupta S, Lee Y, Tao JQ, Akolia I, Louneva N, Forrest K, Paul O, Brooks TG, Grant GR, Sehgal A, Chatterjee S. J Biol Rhythms. 2025 September.
- Generating correlated data for omics simulation Yang J, Grant GR, Brooks TG. PLOS Comp Bio. 2025 September.
- Sampling spiked Wishart eigenvalues Brooks TG. Communications in Statistics - Simulation and Computation. 2025 April.
- Sleep disorders as risk factors for calcific aortic stenosis El Jamal N, Brooks TG, Skarke C, FitzGerald GA. American Journal of Preventive Cardiology 2025 March.
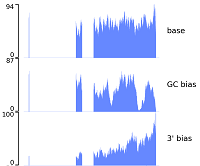
- Sources of non-uniform coverage in short-read RNA-Seq data Brooks TG, Lahens NF, Mrcela A, Yang J, Purohit S, Naik A, Ricciotti E, Sengupta S, Choi PS, Grant GR. bioRxiv. 2025 January.
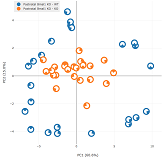
- Genetic variants associated with chronic fatigue syndrome predict population-level fatigue severity and actigraphic measurements Liu PZ, Raizen DM, Skarke C, Brooks TG*, Anafi RC*. Sleep. 2025 February.
- 3-manifolds with constant Ricci eigenvalues (λ, λ, 0) Brooks TG. Geometriae Dedicata. 2024 December.
- BEERS2: RNA-Seq simulation through high fidelity in silico modeling Brooks TG, Lahens NF, Mrcela A, Sarantopoulou D, Nayak S, Naik A, Sengupta S, Choi PS, Grant GR. Briefings in Bioinformatics. 2024 April.
- Challenges and best practices in omics benchmarking Brooks TG, Lahens NF, Mrcela A, Grant GR. Nature Reviews Genetics. 2024 January. https://doi.org/10.1038/s41576-023-00679-6
- Prognostic utility of rhythmic components in 24-h ambulatory blood pressure monitoring for the risk stratification of chronic kidney disease patients with cardiovascular co-morbidity El Jamal N, Brooks TG, Cohen J, Townsend RR, Rodriguez de Sosa G, Shah V, Chronic Renal Insufficiency Cohort Study (CRIC) Consortium, Nelson RG, Drawz PE, Rao P, Bhat Z, Chang A, Yang W, FitzGerald GA, Skarke C. Journal of Human Hypertension. 2024 January. https://doi.org/10.1038/s41371-023-00884-0
- Deep Phenotyping of the Lipidomic Response in COVID-19 and non-COVID-19 Sepsis. Meng H, Sengupta A, Ricciotti E, Mrcela A, Mathew D, Mazaleuskaya LL, Ghosh S, Brooks TG, Turner AP, Schanoski AS, Lahens NF, Tan AW, Woolfork A, Grant GR, Susztak K, Letizia AG, Sealfon SC, Wherry EJ, Laudanski K, Weljie AM, Meyer NJ, FitzGerald GA. Clinical and Translational Medicine. 2023 November. DOI: 10.1002/ctm2.1440
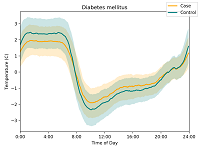
- Diurnal rhythms of wrist temperature are associated with future disease risk in the UK Biobank Brooks TG, Lahens NF, Grant GR, Sheline YI, FitzGerald GA, Skarke C. Nature Communications. 2023 August
- Circadian regulation of lung repair and regeneration Naik A, Forrest KM, Paul O, Issah Y, Valekunja UK, Tang SY, Akhilesh RB, Hennessy EJ, Brooks TG, Chaudhry F, Babu A, Morley M, Zepp JA, Grant GR, FitzGerald GA, Sehgal A, Worthen GS, Frank DB, Morrisey EE, Sengupta S. JCI Insight. 2023 July
- Meta-analysis of diurnal transcriptomics in mouse liver reveals low repeatability of rhythm analyses Brooks TG, Manjrekar A, Mrcela A, Grant GR. J Biol Rhythms. 2023 June
- Sexual dimorphism in the response to chronic circadian misalignment on a high fat diet. Anderson ST, Meng H, Brooks TG, et al. Sci Transl Med. 2023 May DOI: 10.1126/scitranslmed.abo2022
- No increase in inflammation in late-life major depression screened to exclude physical illness. Luning Prak ET, Brooks TG, Makhoul W, Beer JC, Zhao L, Girelli T, Skarke C, Sheline YI. Transl Psychiatry. 2022 Mar 12(1):118. doi: 10.1038/s41398-022-01883-4. PubMed PMID: 35332134.
- Nitecap: An Exploratory Circadian Analysis Web Application. Brooks TG, Mrcela A, Lahens NF, Paschos GK, Grosser T, Skarke CC, FitzGerald GA, Grant GR. J Biol Rhythms. 2022 Feb 37(1):43-52. doi: 10.1177/07487304211054408. Epub 2021 Nov 2. PubMed PMID: 34724846.
- CAMPAREE: a robust and configurable RNA expression simulator. Lahens* NF, Brooks* TG, Sarantopoulou D, Nayak S, Lawrence C, Mrcela A, Srinivasan A, Schug J, Hogenesch JB, Barash Y, Grant GR. BMC Genomics. 2021 Sep 22(1):692. doi: 10.1186/s12864-021-07934-2. PubMed PMID: 34563123; PubMed Central PMCID: PMC8467241.
- Comparative evaluation of full-length isoform quantification from RNA-Seq. Sarantopoulou* D, Brooks* TG, Nayak S, Mrcela A, Lahens NF, Grant GR. BMC Bioinformatics. 2021 May 22(1):266. PubMed PMID: 34034652.
- Loss of circadian protection against influenza infection in adult mice exposed to hyperoxia as neonates. Issah Y, Naik A, Tang SY, Forrest K, Brooks TG, Lahens N, Theken KN, Mermigos M, Sehgal A, Worthen GS, FitzGerald GA, Sengupta S. Elife. 2021 Mar 10. doi: 10.7554/eLife.61241. PubMed PMID: 33650487.
- Accounting for Time: Circadian Rhythms in the Time of COVID-19. Sengupta S, Brooks TG, Grant GR, FitzGerald GA. J Biol Rhythms. 2021 February. 36(1):4-8. doi: 10.1177/0748730420953335. Epub 2020 Sep 2. Review. PubMed PMID: 32875944.
- Nonnegative curvature and conullity of the curvature tensor. Brooks TG. Annals of Global Analysis and Geometry. 2019 August. doi: https://doi.org/10.1007/s10455-019-09678-5.
- Targeting MRTF/SRF in CAP2-dependent dilated cardiomyopathy delays disease onset. Xiong Y, Bedi K, Berritt S, Attipoe BK, Brooks TG, Wang K, Margulies KB, Field J. JCI Insight. 2019 March. 4(6). doi: 10.1172/jci.insight.124629. eCollection 2019 Mar 21. PubMed PMID: 30762586.
* indicates equal contributions